- Principles and applications of polymerase chain reaction in medical diagnostic fields: a review - (http://www.ncbi.nlm.nih.gov/pmc/articles/pmc3768498/)
- Polymerase Chain Reaction (PCR) - (http://www.ncbi.nlm.nih.gov/probe/docs/techpcr/)
- Polymerase Chain Reaction (Interactive) - (https://www.dnalc.org/resources/animations/pcr.html)
- Polymerase Chain Reaction (PCR) Fact Sheet - (https://www.genome.gov/10000207)
What is a PCR?
PCR or Polymerase Chain Reaction is a revolutionary method that is used to amplify specific segments of DNA or RNA. This method was developed by Kary Mullis in the 1980s. Kary Mullis received the Nobel Prize in Chemistry in 1993, for his work on PCR.
PCR aids in increasing copies of genetic material that can be used for further analysis. This procedure allows genetic analysis from small samples of blood and tissue.

PCR amplification of DNA is used routinely in laboratories across the world, as it is fast and inexpensive.
How Does PCR Amplification Occur?
PCR amplification occurs through a series of steps using primers. Primers are laboratory synthesized nucleotide sequences that bind to the complementary strands of DNA. They are the starting points for synthesis and elongation. Primers are designed to include the regions of DNA that need to be amplified by the PCR technique.
Denaturation of DNA:
The sample DNA is heated to 94-960C for a few minutes to denature the DNA and to separate the two strands of DNA.
Anneal Primers:
The temperature is then cooled down instantly to 50-650C to facilitate the attachment of the DNA primers to the complementary strands of DNA.
Once the DNA primers are attached to the complementary strands of DNA from the sample, DNA strand elongation is ready to begin.
Strand Elongation:
In this stage, there is strand elongation which occurs at a temperature of 720C. The enzyme Taq polymerase mediates this process by attaching nucleotides to the DNA primers based on the nucleotides present in the DNA strand of the sample.
On completing this step, an identical copy of the original DNA strand will be obtained.
The steps from DNA denaturation till strand elongation are repeated to get multiple copies of the original DNA strand. The process repeats several times till a billion copies are generated within a few hours.
DNA Copies in Each PCR Cycle
| Cycles in a PCR | No of targeted DNA copies |
| 1 | 2 |
| 2 | 4 |
| 3 | 8 |
| 4 | 16 |
| 5 | 32 |
| 6 | 64 |
| 7 | 128 |
| 8 | 256 |
| 9 | 512 |
| 10 | 1024 |
| 11 | 32,768 |
| 12 | 1,048,576 |
What are Added to the Reaction Mixture to be Loaded into a PCR for DNA Amplification?
The following are added to the PCR reaction mixture for DNA amplification:
- Template DNA
- Primer mix
- Taq polymerase
- Nucleotides
- Buffer

What are the Advances That Have Allowed DNA Amplification to be Automated Using a PCR Machine?
A. DNA polymerases that are thermostable: In Vivo enzymes are used to separate the complementary strands of DNA that are to be amplified. In a PCR, heat is used to denature complementary strands, Taq polymerase was isolated from Thermus aquaticus, an organism that can withstand high temperatures. This allowed DNA amplification using a PCR machine.
B. Altering temperature bath: Thermal cyclers that alternate between varying temperatures allow automation of DNA amplification.
What is the Plateau Effect in a PCR?
During the last stages of a PCR, when the concentration of the product is between 0.3nM to 1.0nM, there is an arrest in the exponential rate of DNA amplification. This could be due to:
- Degradation of nucleotides
- Depletion of reactants
- Competition for reactants
What are the Different Types of PCR?
The different types of PCR are:
- Reverse Transcription (Rt) PCR: This type of PCR is called Rt-PCR as a complementary DNA strand is initially developed from a sample RNA using the enzyme reverse transcriptase (Rt). Rt- PCR is used to study the expression levels of a specific RNA.
- Quantitative PCR (qPCR) or Real Time PCR: This PCR method is used to assess the relative amount of DNA present in a sample using proportions present after amplification.
- Colony PCR: In this type of PCR, colonies of bacteria are added. The initial denaturation step is extended for a longer period to promote cell disruption.
- Hot Start PCR: During room temperature, Taq polymerase may lead to annealing of non-specific primers that result in the elongation of non-specific products. To avoid this, an essential component of the reaction mixture like nucleotides or magnesium ions are added after the initial step, when the temperature is increased to 960C.
This may be carried out manually or by placing wax in between the reaction mixture and the specific component. As the temperature increases, the wax melts and the component is mixed with the reaction mixture.
- Nested PCR: This method of PCR has been designed to reduce non-specific amplification of DNA strands. Initially, an ‘outer’ primer is added that covers a larger length of the DNA strand. After amplification, an ‘inner’ primer is added, that amplifies the shorter, more specific region.
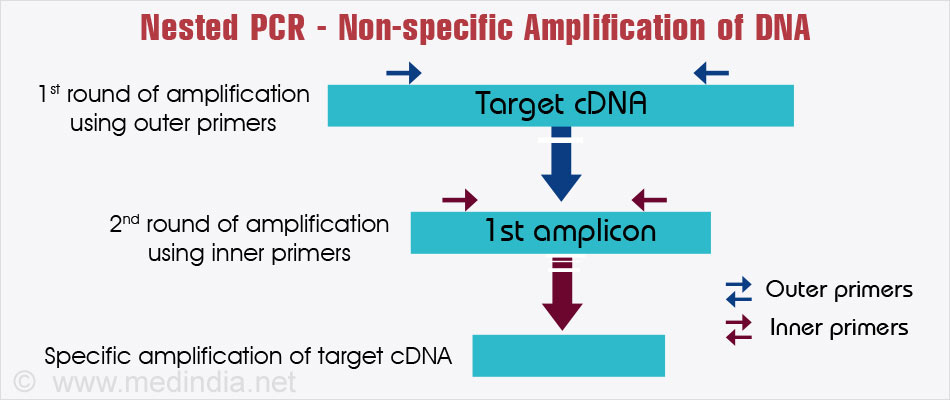
- Touchdown PCR: In this procedure, the initial denaturation temperature is just below the temperature at which the primer melts. Over subsequent cycles, the denaturation temperature is lowered. This method of PCR is used to avoid non-specific primer annealing.
- In-Situ or Slide PCR: The sample is placed on a slide and the reaction mixture is placed over that. This is then covered using a cover slip and the slide placed in a PCR for elongation.
- Inverse PCR: The primers are oriented to elongate DNA sequence in the reverse direction when compared to the standard PCR.
- Multiplex PCR: Multiple primers are used to sequence multiple target DNAs in the same reaction mixture.
- Assembly PCR: Small fragments of DNA are used to generate large DNA oligonucleotides. This procedure can be used to produce synthetic genes.
What is the Difference Between DNA Replication In Vivo and Strand Amplification in a PCR?
DNA replication in vivo or within the living system is different from strand amplification in a PCR in some ways.
- The separation of the complementary strands in preparation for strand elongation takes place using the enzyme DNA helicase invivo while in a PCR the strands are separated using heat denaturation.
- The DNA replication fork (area where the replication of DNA takes place) and Okazaki fragments are not formed in PCR amplification of DNA and strand elongation is continuous.
- The primer for DNA replication is RNA while for PCR amplification, it is DNA.
What are the Uses of Amplifying DNA Using a PCR?
The uses of DNA amplified by PCR are:
- Identification of infections: Amplification of the DNA of viruses, bacteria and other microbes aid in easier identification of pathogens. This in turn aids in providing the right medical treatment. A classic example of the use of PCR amplified DNA in identification of diseases was during the severe acute respiratory syndrome (SARS) epidemic.
- Early identification of cancers: Cancers are formed when there are small mutations in genes. Amplification of DNA can be used to detect cancer causing mutations, even before symptoms arise.
- Organ donation- tissue matching: Tissue matching is an integral part of arranging donors for organ donation and transplantation. Accurate matching aids in lowering risk of tissue rejection by the host.
- Forensic medicine: DNA from a single hair or a small sample of blood can be amplified using a PCR and then used in DNA fingerprinting to identify the individual under study. PCR amplification of DNA is used in forensic medicine to identify victims or suspects and also for paternity testing.
- Evolutionary studies: Since even very small quantities of DNA from fossils can be amplified, PCR is used in evolutionary studies to study the similarities and the differences in genes between organisms.
Limitations of PCR
Some of the limitations of PCR in gene synthesis are as follows:
- Amplicon Size: PCR efficiency decreases with increased amplicon size. Amplicon refers to the DNA fragments or sequences that are amplified by the PCR. PCR has the ability to amplify DNA sequences of up to 3 kb, but ideally the length should be less than 1 kb. The reason is due to the low processivity of the Taq polymerase enzyme which lacks proof reading ability. Many genes, in particular human genes are longer than those that can be multiplied by PCR. Megabases long DNA sequences have to be isolated and then multiplied which is impossible to achieve by PCR.
- Error Rate During Amplification: DNA polymerases have a proof reading mechanism by which they correct the errors committed during DNA replication. Taq polymerase lacks proof reading mechanism and is unable to correct the errors that happen during DNA amplification. The error rate of Taq polymerase is 1 error per 9000 nucleotides.
- Contamination: PCR technique is extremely sensitive and prone to erroneous results due to contaminated DNA. Contaminated DNA may originate from contaminant organisms found in the biological source, airborne cellular debris, products of previous PCR reactions. Contamination can be prevented by using good laboratory technique and taking adequate control measures.