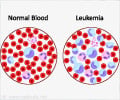
Research has identified a fusion gene responsible for almost 30 percent of of a rare subtype of childhood leukemia with an extremely poor prognosis.

Investigators traced the genetic misstep to the rearrangement of chromosome 16, which brings together pieces of two genes and sets the stage for production of an abnormal protein. The fusion protein features the front end of CBFA2T3, a blood protein, and the back of GLIS2, a protein that is normally produced only in the kidney. Work that appears in the November 13 edition of the journal Cancer Cell reports that in a variety of laboratory models the CBFA2T3-GLIS2 protein switched on genes that drive immature blood cells to keep dividing long after normal cells had died. This alteration directly contributes to leukemia.
AMKL patients with the fusion gene were also found to be at high risk of failing therapy. Researchers checked long-term survival of 40 AMKL patients treated at multiple medical centers around the world and found about 28 percent of patients with the fusion gene became long-term survivors, compared to 42 percent for patients without CBFA2T3-GLIS2. Overall long-term survival for pediatric AML patients in the U.S. is now 71 percent.
"The discovery of the CBFA2T3-GLIS2 fusion gene in a subset of patients with AMKL paves the way for improved diagnostic testing, better risk stratification to help guide treatment and more effective therapeutic interventions for this aggressive childhood cancer," said James Downing, M.D., St. Jude scientific director and the paper's corresponding author. The first author is Tanja Gruber, M.D., Ph.D., an assistant member in the St. Jude Department of Oncology.
Co-author Richard Wilson, Ph.D., director of The Genome Institute at Washington University School of Medicine in St. Louis, noted: "We identified this unusual gene fusion by comparing the genome of children's healthy cells with the genome of their cancer cells. This type of in-depth exploration and analysis is crucial to finding unexpected structural rearrangements in the DNA that can lead to cancer. With this discovery, we now can search for more effective treatment options that target this precise defect."
The study is part of the Pediatric Cancer Genome Project, a three-year collaboration between St. Jude and Washington University to sequence the complete normal and cancer genomes of 600 children and adolescents with some of the most aggressive and least understood cancers. The human genome is the instruction book for assembling and sustaining a person. The instructions are packaged in the DNA molecule. Sequencing the genome involves determining the exact order of the four chemical bases that make up DNA. Human DNA is organized into 46 chromosomes.
Advertisement
When researchers in this study sequenced just the genes that were switched on in the AMKL cells of 14 young patients, the scientists discovered half carried the CBFA2T3-GLIS2 fusion. Additional fusion genes were identified in five of the other patients. Each of those fusion genes occurred in a single patient. The genes involved included HOXA9 and MN1, both previously linked to leukemia, and GATA2 and FLII, which play roles in normal development of the megakaryocytic blood cells that are targeted in AMKL. Megakaryocytes produce the platelets that help blood clot.
Advertisement
"Whole genome sequencing has allowed us to detect alterations in cancer cells that were previously unknown. Many of these changes contribute directly to the development of cancer," Gruber said. "Such sequencing also provides the deeper understanding of the disease that is critical for developing more effective, less-toxic targeted therapies."
GLIS2 is a transcription factor, meaning it attaches to DNA and turns genes on or off. GLIS2 is normally switched off in blood cells and has not been previously linked to cancer.
Working in several laboratory models, researchers showed that GLIS2, either alone or in the fusion gene, increased the activity of other genes in pathways that control cell functions disrupted in cancer. The genes include BMP2 and BMP4, which are now the focus of additional research. The genes are in a pathway that is active early in the developing blood system. This study implicated the genes in AMKL.
Source-Eurekalert