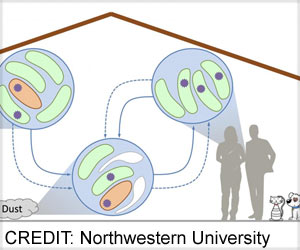
Antibiotic resistance evolves rapidly in patients colonized by diverse bacterial populations, demonstrating a clear link between host diversity and antibiotic resistance.
- Antibiotic resistance poses a global health threat, but the role of host factors remains unknown
- This problem needs to be addressed because resistant infections cause worse outcomes
- Now, it is shown that resistance will evolve rapidly in hosts colonized by diverse pathogen populations
A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance
Go to source).
The evolutionary biology-based concept is that the additional source of standing genetic variation in mixed-strain populations accelerates the evolutionary response to antibiotic treatment by increasing genetic diversity. This implies that antibiotic resistance will evolve rapidly in hosts colonized by diverse pathogen populations.
A new study published in the journal Nature Communications (2✔ ✔Trusted Source
Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients
Go to source) tested the hypothesis that antibiotic resistance evolves rapidly in diverse populations of Pseudomonas aeruginosa, an opportunistic pathogen that causes hospital-acquired infection, particularly in immunocompromised and critically ill patients.
Previous studies have shown that different strains of Pseudomonas can co-occur in the lungs of patients who suffer from chronic P. aeruginosa infection associated with cystic fibrosis and bronchiectasis.
Researchers tested the impact of within-patient Pseudomonas diversity in ICU patients using samples that were collected from ASPIRE-ICU, an observational trial of Pseudomonas infection in European hospitals.
Hidden Threat Within Body: How Pathogen Diversity Drives Antibiotic Response
To characterize the diversity of P. aeruginosa within patients we sequenced the genomes of 441 isolates that were collected from lower respiratory tract samples of 35 ICU patients from 12 hospitals.Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients
Go to source).
They also found evidence of gene mutations that have previously been implicated in resistance, and an outstanding challenge is to determine if these changes were present as standing genetic variation at the onset of treatment or if these changes arose as a result of spontaneous mutations during antibiotic treatment.
The deal between resistance and growth rate makes it challenging to understand how strains that vary in resistance can coexist within the same patient for the long term. Therefore, researchers speculate that host diversity will be high in settings with a high infection rate or in patients where antibiotic exposure is variable across host tissues (3✔ ✔Trusted Source
Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute Pseudomonas aeruginosa infection
Go to source).
This study underscores the importance of within-host bacterial diversity for understanding antibiotic resistance. Using bacterial isolates to measure within-host diversity limited the number of patients included in this study.
Hence, measuring the diversity of pathogen populations might make it possible to predict the likelihood of treatment failure more accurately for individual patients, in the same way, the measurements of diversity in cancer cells have been informative for predicting the success of cancer treatment.
References:
- A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance - (https://bmcinfectdis.biomedcentral.com/articles/10.1186/1471-2334-14-13)
- Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients - (https://www.nature.com/articles/s41467-023-39416-2)
- Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute Pseudomonas aeruginosa infection - (https://www.nature.com/articles/s41467-021-22814-9)
Source-Medindia