Rice University bioengineers have examined a portion of the tuberculosis genome that responds to stress and have found a network of genes that may switch the disease into dormancy.

Oleg Igoshin and Abhinav Tiwari examined a network of genes that may make this possible.
A computer model of the network showed it could act as a "persistence" switch that toggles the organism from a fast growing to a slow-growing state.
"The molecular mechanisms that allow Mycobacterium tuberculosis to switch into this slow-growing, persistent state have been associated with genes that are activated when the microorganism is under stress," said Igoshin.
Tiwari, lead author of the study and a graduate student in Igoshin's lab, said: "We examined a stress-response network of genes that are found in both the TB bacterium and other closely related mycobacteria. We analyzed the role of multiple feedback loops in this network, and were eventually able to identify an ultrasensitive mechanism that works in combination with the feedback loops to form a switch. This switch can possibly activate transition to the persistent state."
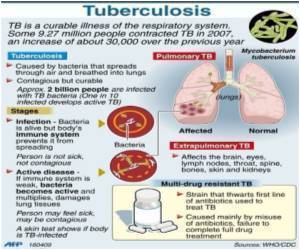
For a long time it's known that the TB bacterium has the ability to "hunker down" and go dormant under stressful conditions.
Advertisement
Igoshin said advances in molecular microbiology have allowed researchers to identify networks of mycobacterial genes that become activated when the organism is stressed.
Advertisement
"Our collaborative team developed an approach that allowed us to formulate general conclusions about the properties of the mycobacterial stress-response network, even though we had limited knowledge of the underlying parameter values," said Igoshin.
Tiwari said, "Using this approach, we systematically examined the different modules, or subsets, of the full network. We found that bistability was linked to a positive feedback loop between MprA and SigE, a protein that binds to RNA polymerase to promote the production of both MprA and SigE."
The researchers believe their modular approach to investigate the role of multiple feedback loops could also be used to unravel mechanisms that other bacteria use to control bistability.
"There are many outstanding questions regarding the specific ways that gene regulatory networks operate in bacteria. The generality of this modular approach opens up a promising avenue for answering some of those questions because it can be readily adapted to other networks," said Igoshin.
The study has been published in Physical Biology.
Source-ANI