A new study led by Anhui Medical University and BGI suggests that coding variants in immune disease-related genes play only a small part in the overall genetic risk for psoriasis.
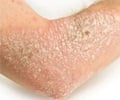
Psoriasis is a complex, chronic, lifelong skin disease. It typically first strikes people between the ages of 15 to 35, but can affect anyone at any age, including children. This terrible disease is the results of the interaction of multiple factors, such as environment, genetics, and immunology. The rapid and cost-effective sequencing technologies have enabled researchers to dig out numerous risk-associated variants in psoriasis, but the functional coding variants, particularly low-frequency and rare variants, have not been systematically investigated.
In this study, researchers took two-phase to identify coding variants. In the discovery stage, they conducted exome sequencing on 781 patients with psoriasis and 676 people without psoriasis as control. The efforts yielded 518,308 single-nucleotide variants (SNVs). Of these variants, 20.62% were nonsynonymous, and 68.13% were rare.
Considering the limitation of sample size and techniques in the discovery stage, researchers performed 2 independent studies in a large sample of 9,946 patients with psoriasis and 9,906 controls using targeted sequencing. A total of 3.2 Mb of coding DNA surrounding the targeted regions of 1,326 genes (covering 133 SNVs, 622 immune disease–related genes, and some top genes) was captured. They totally identified 82,387 nonsynonymous SNVs, of which 97.07% were rare.
Through further analysis, they discovered two independent missense SNVs in IL23R and GJB2 with low frequency and five common missense SNVs in LCE3D, ERAP1, CARD14 and ZNF816A associated with psoriasis at genome-wide significance. The rare missense SNVs in FUT2 and TARBP1 were also observed with suggestive evidence of association with this disease.
In addition to the SNVs analysis, researchers investigated 622 immune disease-related genes, and the results showed that the coding variants, at least common and low-frequency nonsynonymous variants, have limited independent contribution to psoriasis risk. Taking all the findings together, the study indicated that nonsynonymous SNVs in the 1,326 targeted genes had limited contribution to the overall genetic risk of psoriasis.
Advertisement
Advertisement
Source-Eurekalert