Researchers at Johns Hopkins and the Karolinska Institutet say their study bridges the gap between whole-genome genetic sequencing and diseases that have no single or direct genetic cause.
The research was published in Nature Biotechnology Jan. 20. Most genetic changes associated with disease do not occur in protein-coding regions of DNA, but in their regulatory regions, explains Andrew Feinberg, M.D., M.P.H., a Gilman scholar, professor of molecular medicine and director of the Center for Epigenetics at the Johns Hopkins University School of Medicine's Institute for Basic Biomedical Sciences. "Our study analyzed both and shows how genetics and epigenetics can work together to cause disease," he says.
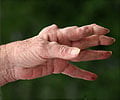
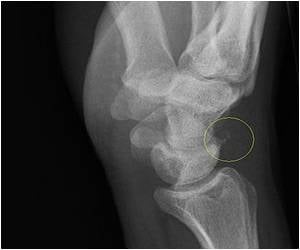
Rheumatoid arthritis is a debilitating disease that causes inflammation, stiffness, pain and disfigurement in joints, especially the small joints of the hands and feet. It is thought to be an autoimmune disease, meaning that the body's immune system attacks its own tissues, an assault led primarily by white blood cells. According to Feinberg, several DNA mutations are known to confer risk for RA, but there seem to be additional factors that suppress or enhance that risk. One probable factor involves chemical "tags" that attach to DNA sequences, part of a so-called epigenetic system that helps regulate when and how DNA sequences are "read," how they're used to create proteins and how they affect the onset or progress of disease.
To complicate matters, Feinberg notes, the attachment of the tags to particular DNA sequences can itself be regulated by genes. "The details of what causes a particular sequence to be tagged are unclear, but it seems that some tagging events depend on certain DNA sequences. In other words, those tagging events are under genetic control," he says. Other tagging events, however, seem to depend on cellular processes and environmental changes, some of which could be the result, rather than the cause, of disease.
To tease apart these two types of tagging events, the researchers catalogued DNA sequences and their tagging patterns in the white blood cells of more than 300 people with and without one form of RA.
The team then began filtering out the tags that did not appear to affect RA risk. For example, if tags were seen on the same DNA sequence in those with and without RA, it was assumed that the tags at those sites were irrelevant to the cause or development of the disease. Then, from among the RA-relevant tags, they narrowed in on tags whose placement seemed to be dependent on DNA sequence. Finally, they made sure that the DNA sequences identified were themselves more prevalent in patients with RA. In this way, they created a list of DNA sequences associated with altered DNA tagging patterns, both of which were associated with RA.
Advertisement
"Our method allows us to predict which tagging sites are most important in the development of a disease. In this study, we looked for tagging sites under genetic control, but similar tags can be triggered by environmental exposures, like smoking, so there are many applications for this type of work," says Yun Liu, Ph.D., a lead researcher on the project.
Advertisement
Source-Eurekalert