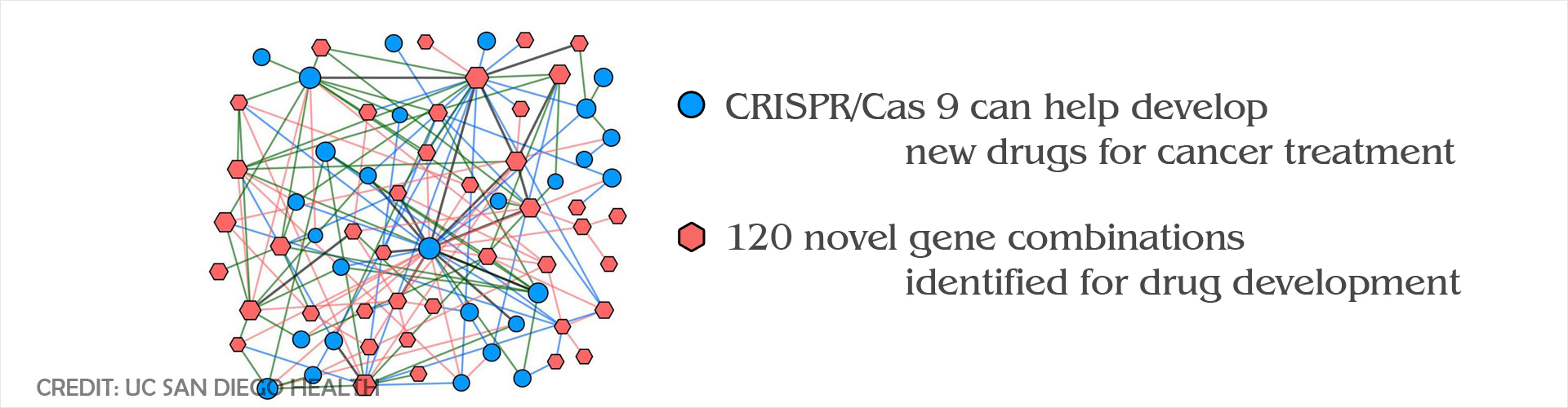
CRISPR gene editing tool was used to identify synthetic-lethal genetic interactions which could be used to develop new drug targets for cancer therapy.
- A research team, from Jacob’s School of Engineering, used the genome editing tool CRISPR to identify gene interactions in synthetic or mutated cancer cells
- The scientists identified 120 novel synthetic-lethal gene combinations
- Synthetic lethality is a method for drug therapy whereby potential new drugs can be designed
CRISPR/Cas9 System
The process of identifying gene combinations that are lethal can be laborious; however, this process is reduced by using the gene editing tool CRISPR/Cas9. This popular tool can test for thousands of synthetic-lethal combinations at the same time.Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated system or Cas genes are required for adaptive immunity among certain bacterial species, which allows these organisms to react to the invading genetic material. The gene editing tools were discovered in E. coli in 1980 but their function did not become evident till 2007. It was shown that S. thermophilus could develop a resistance towards infection by a bacteriophage by integrating a fragment of the genome of the infectious agent into its locus.
Types of CRISPR
There are three different types of CRISPR which have been identified among which type 2 is well known. In the type 2 form of CRIPSR- The DNA from the invading plasmids or viruses are cut into smaller fragments
- These fragments are then incorporated into the CRISPR locus in between short repeats
- There is transcription of the loci; the transcripts are then processed to result in small RNAs
- These small RNAs guide the endonucleases which target the DNA of the invading virus or plasmid based on complementarity
- The Cas9 protein is required for gene silencing
- The protein is also required for the synthesis of CRISPR RNA as well as for the destruction of the target DNA
- Targeting the commonly mutated tumor suppressing gene
- Targeting a gene that can be disrupted by a drug for cancer
A professor at the UC San Diego School of Medicine and co-director of the Cancer Cell Map Initiative, Dr. Trey Ideker, who is also the co-author of the study, said that identifying the genetic interactions using the CRISPR/Cas9 system will aid in revealing functional relationships of genes, associated with a protein complex or a pathway. Dr. Trey, also the founder of the UC San Diego Center for Computational Biology and Bioinformatics said that these findings could be used to improve basic understanding of various biological systems and could also be used for developing drug therapy for cancer.
Synthetic-lethal Interactions
The synthetic-lethal interactions that were identified were found in just one of the three cell lines that were tested, indicating that the interactions were different for the different types of cancer. This is a critical understanding for drug development.CRISPR in Cancer Research
Dr. Si Chen and colleagues published a paper in the International Journal of Biological Sciences titled “CRISPR-Cas9: from Genome Editing to Cancer Research”. The development of cancer involves multiple steps and is associated with innate mutations as well as mutations that are acquired. These mutations lead to the functional abnormality seen in cancer and they determine the progress of cancer. The CRISPR-associated 9 (CRISPR-Cas9) system provides a genome editing approach which is convenient.- Loss of function of gene-screening invivo
- Gain of function of gene-screening invivo
- Acceleration of cancer in animal models for research purposes
- Potential use in gene therapy for cancer
References:
- CRISPR-Cas9: from Genome Editing to Cancer Research - (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5166485/)