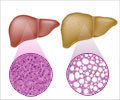
Analysis of microorganisms found in the fecal matter and gut of patients with non-alcoholic fatty liver disease, could predict whether they are at a greater risk for developing cirrhosis that causes irreversible scarring of the liver.
‘Accurate stool microbiome-based non-invasive test could be used to identify patients who are at a greater risk for developing cirrhosis. This test can be used clinically as a diagnostic tool to avoid the deadly and progressive complications of non-alcoholic fatty liver disease.’
Read More..




"The findings represent the possibility of creating an accurate, stool microbiome-based, non-invasive test to identify patients at greatest risk for cirrhosis," said senior author Rohit Loomba, MD, professor of medicine in the Division of Gastroenterology at UC San Diego School of Medicine and director of its NAFLD Research Center. "Such a diagnostic tool is urgently needed."Read More..
Loomba said a novel aspect of the study is the external validation of gut microbiome signatures of cirrhosis in participant cohorts from China and Italy. "This is one of the first studies to show such a robust external validation of a gut microbiome-based signature across ethnicities and geographically distinct cohorts.
The work builds upon previous published research in 2017 and 2019 by Loomba and colleagues.
A link between NAFLD and the gut microbiome is well-documented, but specifics were scant and it has not been clear that discrete metagenomics and metabolomics signatures might be used to detect and predict cirrhosis. In the latest study, researchers compared the stool microbiomes of 163 participants encompassing patients with NAFLD-cirrhosis, their first-degree relatives and control-patients without NAFLD.
Combining metagenomics signatures with participants' ages and serum albumin (an abundant blood protein produced in the liver) levels, the scientists were able to accurately distinguish cirrhosis in participants differing by cause of disease and geography.
Advertisement
Source-Eurekalert