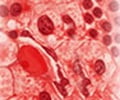
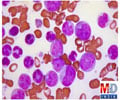
New study provides fresh insights into why some with age-related clonal hematopoiesis, or ARCH go on to develop acute myeloid leukemia (AML) and others don’t.
‘Discovery could provide early warning for blood malignancies, triggering proactive screening and enabling early treatment.’





The study, co-led by Dr. Philip Awadalla, Senior Principal Investigator and Director, Computational Biology at the Ontario Institute for Cancer Research (OICR) and Dr. Quaid Morris, Member, Computational and Systems Biology, Memorial Sloan Kettering Cancer Center (MSK) and OICR Associate, shows how the interplay of positive, neutral and negative evolutionary selection acting on mutations in aging blood stem cells can lead to AML in some individuals with ARCH. They did so by illustrating how negative selection, or ‘purifying selection’, present in individuals who did not go on to develop a malignancy, prevents disease-related cells from coming to dominate the cell population. These discoveries allow for the differentiation between those with ARCH who are at increased risk of developing AML and those who are not. “We have shown that the constellation of evolutionary forces at play within hematopoietic stem cells can be a robust indicator of those who are at increased risk of blood cancers such as AML,” says Awadalla. “Being able to accurately classify patients based on risk can allow for more frequent and intensive screening for those with ARCH mutations with a concerning evolutionary signature.”
The research team computationally generated more than five million blood populations, trained a deep neural network model (a type of machine learning) to recognize different evolutionary dynamics and employed the model to analyze blood samples that had undergone deep genomic sequencing. These samples were from 92 individuals who went on to develop AML, and 385 who did not despite the presence of ARCH. The study is one of the first to use a single system of tools to capture the interaction of the multiple evolutionary forces at play in ARCH.
“The models we developed in this study can significantly increase the value of ARCH as a biomarker for blood malignancies,” says Morris. “Our team is looking forward to continuing to bolster our understanding of ARCH and seeing these advancements help patients.”
The researchers were able to show that these alternative evolutionary models were predictive of AML risk over time. Similarly, these tools were able to identify genes where mutations that are damaging to stem cells can accumulate.
Advertisement
“In the future, we can anticipate screening blood samples for early detection of disease and blood cancers. With these tools we can more proactively monitor people’s health. Early detection of cancer is critical with respect to prevention and effectiveness of treatment,” adds Awadalla.
Advertisement