Observing the long-term structural plasticity in individual spines requires substantial time and effort.
‘The newly developed software that automates brain imaging helps increase productivity. The ability to stimulate and image multiple spines in parallel greatly decreases the cost of running these experiments.’





A
new technique, developed by MPFI researchers, automates the process to
make observing and quantifying this growth far more efficient. The
open-source method is available to any scientist hoping to image
plasticity as it happens in dendritic spines using Scanimage. The work
was published in January 2016 in the Public Library of Science journal, PLOS ONE.Scientists working in Ryohei Yasuda's laboratory at MPFI are working to understand how proteins facilitate the plasticity of dendritic spines, the biological basis of learning and memory.
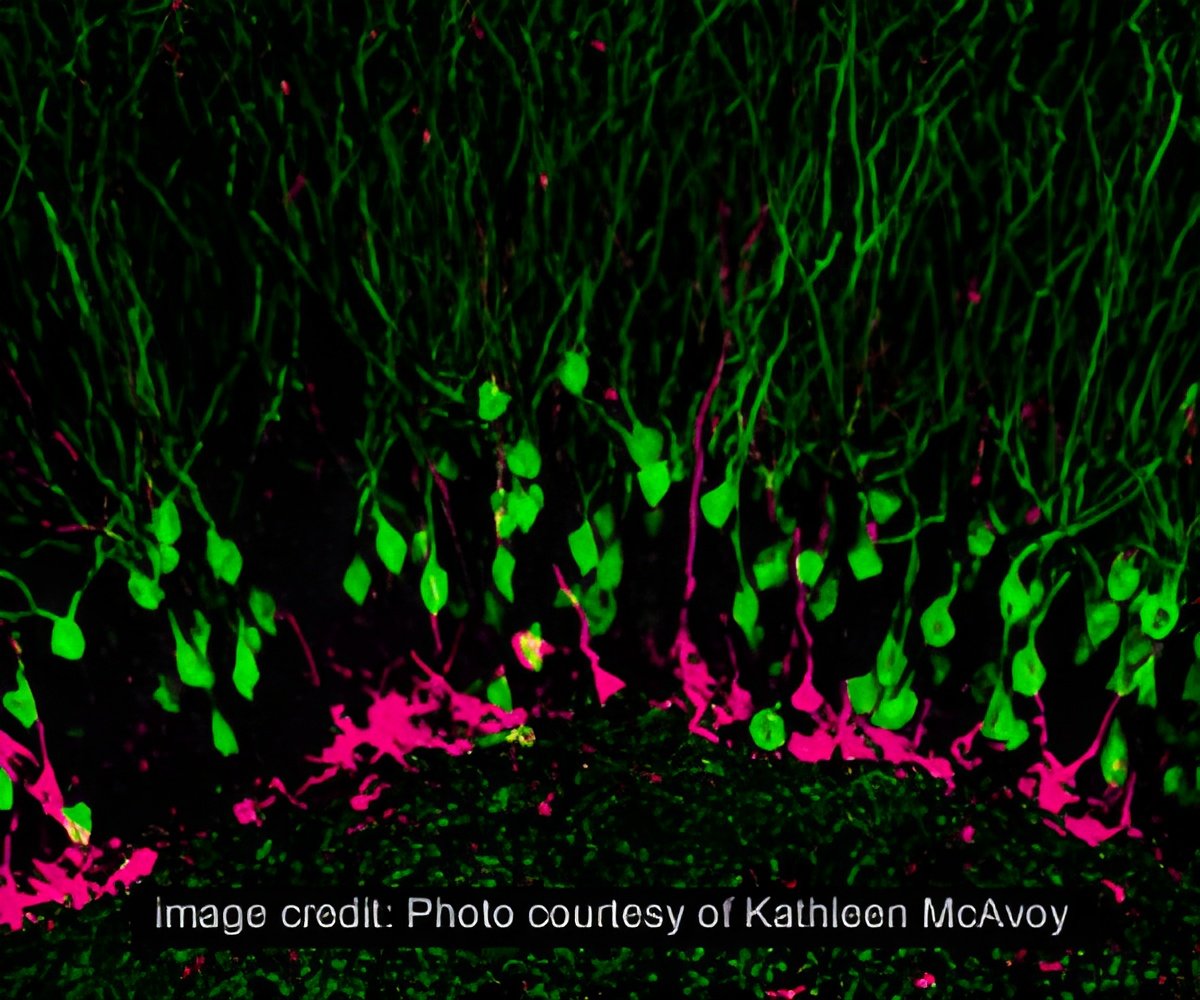
They use two-photon microscopy, an advanced technique for live-cell imaging, and glutamate uncaging, a technique that can induce plasticity in individual spines of interest using light. This is a meticulous process, wherein a scientist must continually focus the microscope on a single dendritic spine over an extended period, often an hour or longer.
Michael Smirnov, Post-doctoral researcher at MPFI, developed a software that allows the computer to automatically track, image, and stimulate up to five dendritic spines at a time. "We can collect the data and figure out the proteins responsible much quicker with this [program] because we can run much more robust experiments," said Smirnov.
In addition to increasing productivity, the ability to stimulate and image multiple spines in parallel greatly decreases the cost of running these experiments.
Advertisement
In contrast to previous open-sourced focusing programs, this one implements a highly capable and customizable focus and drift correction system to ensure that it can be used for a variety of biological applications. "The paper explains further modifications to make the process automated," said Smirnov. "It shares the open source code, so essentially, other people from other institutes can easily pick this up and use it for themselves."
Advertisement