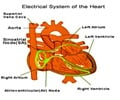
Scientists have developed a supercomputing drug screening for deadly heart arrhythmias. The sudden cardiac arrest causes the most deaths by natural causes in the U.S., estimated at 325,000 per year.
“What we set out to do was to try to solve that problem by building a computer-based pipeline for screening,” said Colleen Clancy, a professor in the Department of Physiology and Membrane Biology and the Department of Pharmacology at the UC Davis School of Medicine. Clancy co-authored a study on the computational cardiotoxicity drug screening pipeline in the journal Circulation Research in April 2020.
“The major novelty of the pipeline is that we found a way to connect the atomistic scale to higher level function scales, like protein function, cell function, and in our simulated tissue-level models we can calculate the spatial and temporal gradients of electrical activity in those simulated pieces of tissue,” Clancy said. “That is an approximation of the electrocardiogram that's measured in the clinic. We can do a direct comparison between the electrocardiogram in the simulated tissue, and electrocardiograms from patients that have taken those drugs.”
Up until the early 2000s, the reason most drugs were removed from the market following FDA approval was cardiotoxicity in the form of deadly arrhythmia. In 2005, the FDA required a separate test for all drugs. It measured the average time between the Q and T waves on an electrocardiogram, a record of the heartbeat. QT prolongation became a red flag for drug cardiotoxicity. But one problem is that some harmless substances, like grapefruit juice, also prolong QT interval, and using it as a proxy for heart arrhythmia could mean the loss of potentially useful and safe drugs.
The two drugs chosen in the study both prolonged the QT interval. One of them, dofetilide, is a known proarrhythmic agent. The other, moxifloxacin, has a strong safety profile in healthy humans. “There's been no way to distinguish between those two classes,” said Clancy. “That's what we were able to show in the computational pipeline.”
Starting from the chemistry of the drug interactions with a target, the scientists used that information to predict proarrhythmia vulnerability through a machine learning approach based on multi-scale computer simulation data.
Advertisement
"The big challenge computationally is the system that we studied is pretty large," said study co-author Igor Vorobyov, an assistant professor in the Department of Physiology and Membrane Biology and the Department of Pharmacology at the UC Davis School of Medicine. “It's on the atomistic scale. We have around 130,000 atoms in our system. This includes the hERG protein embedded in the lipid membrane surrounded in a salt aqueous solution in water.”
Advertisement
Researchers were awarded time on the Stampede2 system at the Texas Advanced Computing Center (TACC), as well as Comet at the San Diego Supercomputer Center (SDSC), making use of Comet's GPU and CPU nodes. The National Center for Supercomputing Applications (NCSA) allocated use of its National Science Foundation-funded Blue Waters system. For long-term (greater than 5 microsecond) simulations of the relatively slow entry of the drug into cardiac cells, the scientists also made use of Anton 2, a special-purpose supercomputer for biomolecular simulation designed and constructed by D. E. Shaw Research (DESRES) and hosted by Pittsburgh Supercomputing Center (PSC).
The team used enhanced sampling simulations, called umbrella sampling, to facilitate the molecular dynamics simulations and yield quantitative determination of the binding affinities and rates of reaction needed for linking scales and feeding parameters further up the pipeline to the functional model.
“That was the novel linkage between our scales that we've both worked on for many years,” added Clancy. “Until now, there was no way to really connect those scales in a meaningful way.” The researchers are taking the next steps in their computational pipeline to expand the set of drugs screened for cardiotoxicity, by looking at a variety of agents that have been part of the Comprehensive In Vitro Proarrhythmia Assay (CiPA) safety pharmacology testing group. They're also working with the biopharmaceutical company Amgen to get blinded compounds for their screening.
“The idea is that Amgen would give us the chemical structures of the drugs, but we wouldn't know whether they were proarrhythmic or not,” said Clancy. “Then we'll use the pipeline to make some predictions about those drugs, and send those results back where they will reveal if our model was accurate in predicting what they've seen.”
Clancy and the team also want to move the research into a personalized medicine approach, building models of an individual's cellular electrophysiology that include some genetic background. One project cultures heart muscle cells from individuals to develop a model called the ‘induced pluripotent stem cell-derived cardiac myocyte.’
"The benefit of a personalized medicine approach is that it includes all of the genetic information in the background, so one can screen drugs and then start to ask why some individuals are susceptible to drugs and proarrhythmia from drugs, while other people aren't,” said Clancy. “We may even be able to work toward developing reconstructions of their hearts in silico.”
In addition to the NSF supercomputer allocations, the study was primarily funded by the National Institutes of Health and the American Heart Association.
About SDSC
The San Diego Supercomputer Center (SDSC) is a leader and pioneer in high-performance and data-intensive computing, providing cyberinfrastructure resources, services, and expertise to the national research community, academia, and industry. Located on the UC San Diego campus, SDSC supports hundreds of multidisciplinary programs spanning a wide variety of domains, from astrophysics and earth sciences to disease research and drug discovery. In late 2020 SDSC will launch its newest National Science Foundation-funded supercomputer, Expanse. At over twice the performance of Comet, Expanse supports SDSC’s theme of ‘Computing without Boundaries’ with a data-centric architecture, public cloud integration, and state-of-the art GPUs for incorporating experimental facilities and edge computing.
Source-Newswise